Estimating Variant Parameters Using Population Statistics
This is a work in progress - please ignore for now
Abstract
Use the two variants Alpha and Delta to construct quantities, the difference in growth rate and the ratio of reproduction numbers, where the behavioural effect cancels out. Focus on quantities like this that cancel out behaviour as this appears to make the series very stable, so amenable to accurate estimation.
Under certain assumptions that are stated below, the growth rates in the Alpha and Delta variants, $$\lambda_t(\alpha)$$ and $$\lambda_t(\delta)$$, appear to satisfy a linear relationship $$A\lambda_t(\alpha)-B\lambda_t(\delta)=1$$ for determinable constants $$A$$ and $$B$$. One possible explanation for this is that $$A$$ and $$B$$ are proportional to the respective generation times of the two variants, though other explanations are possible. If this explanation is correct then it suggests the generation time of Delta is slightly less than that of Alpha. Combining this with external estimates of the transmission advantage of Delta over Alpha leads to separate estimates of the generation times of Alpha and Delta.
Background
In the UK the variant story was Alpha was almost exclusive in Feb and March 2021. Delta entered in April, because equal with Alpha in mid May, and became virtually the exclusive variant by the end of June. So Feb-June 2021 was essentially a two-variant story, which conveniently meant the S-gene was an excellent proxy distinguishing Delta from Alpha.
[Discussion about other uses for the method of cancelling behaviour effects for two or more variants.]
Going to use COG-UK genome sequence counts, mainly because they are continually published and available at day resolution. SGTF counts could also work, but as far as I am aware are not publicly available at day resolution, are not available for all testing laboratories, and ceased publication around 20 June.
For the following discussion, it will be necessary that the sample counts from COG-UK are representative of PCR-confirmed cases during the periods considered, so that the counts of Alpha and Delta are in the correct proportion each day. Appendix 1 attempts to make the case these counts are representative between the dates 20 April and 31 May 2021 and (with a separate argument based on comparing with SGTF counts) that the sequenced counts are likely representative between 20 April and 20 June. Prior to 20 April, genome sequencing may have been preferentially directed towards S-gene positive cases in order to track the then-new B.1.617.* family.
First thing
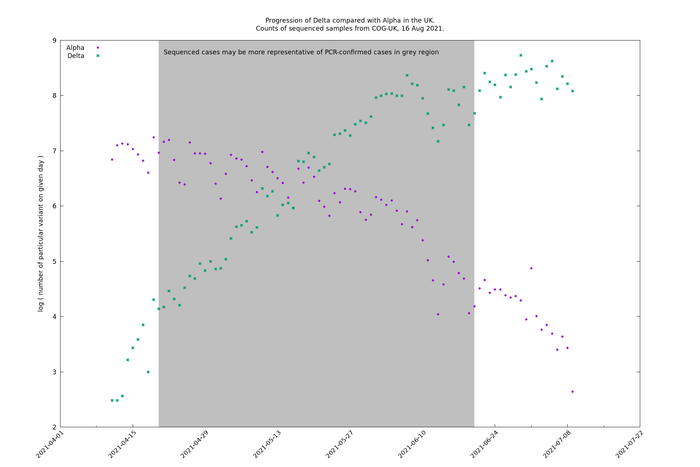
Consider raw counts of Alpha and Delta.
On a log scale, both variants are broadly linear (i.e., constant growth) up to 1 June. Delta deviates from linearity before mid-April which may be due to non-representative genomic sampling, or the effect of travel cases before India was added to the red list on 23 April. After 1 June there is a significant reduction in growth in both variants, which may be due to the heatwave in the first half of June. The points are fairly noisy, which is partly due to day-of-the-week effects.
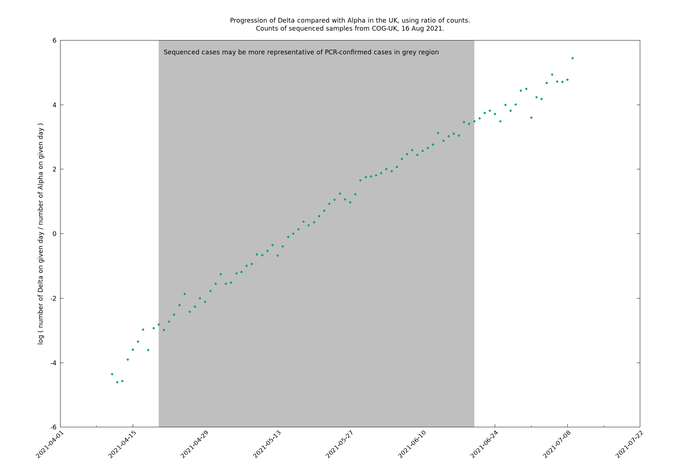
Now let's take the ratio, Delta/Alpha, again on a log scale.
This is a remarkably noiseless and stable curve - striking for real-world data. Note that there is no processing or smoothing involved in this, only restricting to days for which the counts are at least 10. (An alternative presentation would be to include error bars, though that is quite cluttering.)
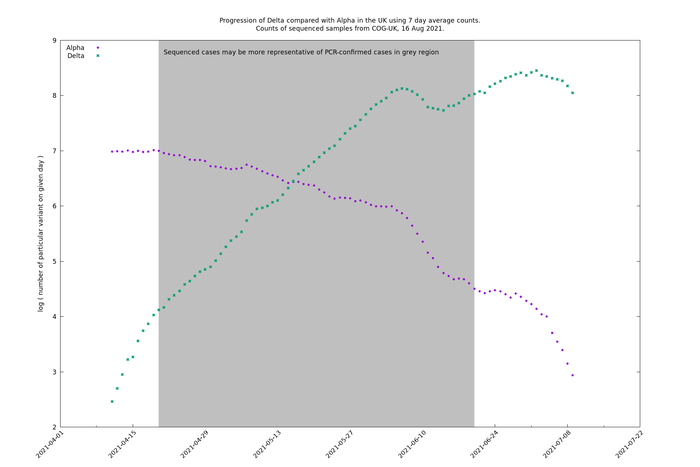
The stability of the Delta/Alpha graph compared with the separate Alpha and Delta graphs may be partly due to the fact that the day-of-the-week effect has been cancelled out, so let's repeat the comparison with 7-day-moving-average (7dma) counts.
Now the (presumably) behavioural effect is much clearer. There was a big change around 1 June that lead to significant changes in the Alpha and Delta trajectories, but the ratio Delta/Alpha remains extremely stable, close to remaining a straight line on a the log scale line, though bending over slightly.
In what follows there will be different analyses of the above phenomenon relying on different sets of assumptions, moving broadly from the more conservative assumptions to the more speculative. Because there are several possible competing effects, it is not possible to completely disentangle them based on these data alone, so we shall analyse one effect at a time, as if it were the main driver of the outcome we see and the other effects are negligible. In reality it may be that the outcome is produced by a combination...[But mention how it turns out that vaccine effect is less likely, so it's not so bad].
Note that all regression is carried out on unsmoothed data, though sometimes the regression lines are plotted on 7dma smoothed data for clarity. (It is possible to regress on to smoothed data, but it would be statistical can of worms to interpret the result because of the strong dependency between neighbouring days.)
Inner time period (2021-04-20 - 2021-05-31) looking for differential vaccine (or other immunity) effect
Running assumptions:
- People's behaviour does not significantly depend on the variant type they are infected with
- Genomic sampling is representative over the period 2021-04-20 - 2021-05-31
Negative binomial regression over this period gives
\[\lambda(\delta)-\lambda(\alpha) = 0.114\;(0.111\; -\; 0.117)\; \text{per day}\]
[Graph of Delta/Alpha with regression line]
The ONS antibody survey shows that antibody positivity in adults rose from 45% to 81% over this period, largely due to vaccination (though party due to infection). If Delta escaped infection+transmission immunity more than Alpha then we would expect to see an upwards bend in the curve (and vice-versa if Alpha escaped more than Delta), but we do not see that, so here we have some indirect evidence that Alpha and Delta do not have very different vaccine escape with respect to infection+transmission immunity.
Ratio $$R_t(\delta)/R_t(\alpha)$$ using secondary attack rate survey
Public Health England provides a secondary attack rate (SAR) survey as a by-product of NHS Test and Trace: see Technical Briefing 20, section 1.6.
This survey provides counts of the number of contacts of each case under study, and also the number of contacts who go on to test positive themselves. These are stratified by the variant type of the case, as determined by sequencing or genotyping. In any given week, the ratio of the SAR of Delta to that of Alpha is an estimate of $$R_t(\delta)/R_t(\alpha)$$, which in turn could be an estimate of $$R_0(\delta)/R_0(\alpha)$$ under the further assumption of equal infection+transmission vaccine escape of Alpha and Delta.
Similarly to the constant growth rate argument in the previous section, there is not an obvious increasing or decreasing trend of $$R_t(\delta)/R_t(\alpha)$$ over the period 26 April - 6 June 2021 during which time overall vaccine immunity increased substantially, so this is indirect evidence that there is not a large difference between Alpha and Delta in infection+transmission vaccine escape (though again it is possible there is such an effect that is mixed up with other effects).
Under the equal infection+transmission vaccine escape assumption, these weekly ratios can be combined, weighted by their certainty, to form an overall estimate of $$R_0(\delta)/R_0(\alpha)$$. This came to 1.650 (1.583 - 1.720). The calculation can be found here.
(Describe method.)
Inner time period (2021-04-20 - 2021-05-31), assume generation times are equal
Running assumptions:
- People's behaviour does not significantly depend on the variant type they are infected with
- Genomic sampling is representative over the period 2021-04-20 - 2021-05-31
- The SAR survey is providing a good estimate of secondary attack rates
- Vaccine escape against infection+transmission is similar for Alpha and Delta
- Generation time is the same for Alpha and Delta, and unchanging over this period
- Generation time can be approximated by a constant distribution (so $$R=e^{\lambda T}$$)
Under the above assumptions we have
- $$\lambda(\delta)-\lambda(\alpha) = 0.114$$ ($$0.111$$ - $$0.117$$) per day,
- $$R_t(\delta)/R_t(\alpha)=1.650$$ ($$1.583$$-$$1.720$$),
- $$R_t(\alpha)=e^{\lambda(\alpha) T}$$, and $$R_t(\delta)=e^{\lambda(\delta) T}$$ for the common generation time $$T$$
which together imply $$T=\log(1.650)/0.114=4.4$$ ($$4.0$$ - $$4.8$$) days.
Inner time period (2021-04-20 - 2021-05-31), different generation times of Alpha and Delta, estimate growth rates separately
Running assumptions:
- People's behaviour does not significantly depend on the variant type they are infected with
- Genomic sampling is representative over the period 2021-04-20 - 2021-05-31
- The SAR survey is providing a good estimate of secondary attack rates
- Vaccine escape against infection+transmission is similar for Alpha and Delta
- Generation times are unchanging over this period
- Generation time can be approximated by a constant distribution (so $$R=e^{\lambda T}$$)
Over this time period, the growth rates of Alpha and Delta are both fairly constant. Negative binomial regression gives $$\lambda(\alpha) = -0.024$$ ($$-0.027$$ - $$-0.021$$) per day, $$\lambda(\delta) = 0.086$$ ($$0.083$$ - $$0.090$$) per day.
Under the above assumptions we have
- $$\lambda(\alpha) = -0.024$$ ($$-0.027$$ - $$-0.021$$) per day,
- $$\lambda(\delta) = 0.086$$ ($$0.083$$ - $$0.090$$) per day,
- $$R_t(\delta)/R_t(\alpha)=1.650$$ ($$1.583$$-$$1.720$$),
- $$R_t(\alpha)=e^{\lambda(\alpha) T(\alpha)}$$, and $$R_t(\delta)=e^{\lambda(\delta) T(\delta)}$$
which together imply $$0.024T(\alpha)+0.086T(\delta)=\log(1.650)\approx 0.501$$ (+ error bars).
Because the Alpha growth is much smaller in magnitude than the Delta growth, the relationship between $$T(\alpha)$$ and $$T(\delta)$$ is relatively insensitive to $$T(\alpha)$$, so if $$T(\alpha)$$ is in the range $$4$$-$$6$$ days, say, then the above relationship would imply $$T(\delta)$$ is in the narrower range $$4.2$$ to $$4.7$$ days using central estimates, or [fill in] using the confidence intervals for $$\lambda(\alpha)$$, $$\lambda(\delta)$$ and $$R_t(\delta)/R_t(\alpha)$$.
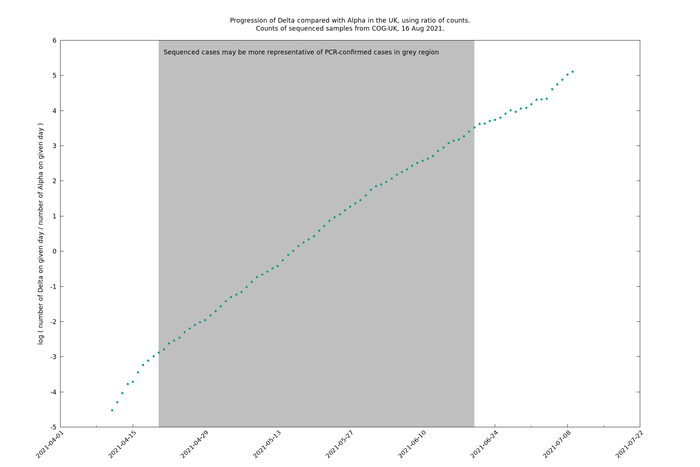
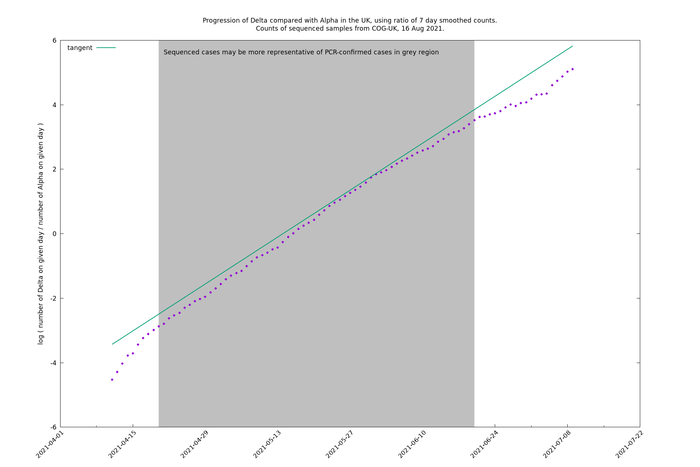
Wider time periods (2021-04-20 - 2021-06-20 or 2021-07-08)
If we extend the time period to 2021-06-20, the latest date for which there is some evidence that the genomic sampling is representative, or to 2021-07-08, the latest date for which there is a non-negligible amount of Alpha, then we see the Delta/Alpha relative count curve bends over downwards (i.e., in a concave manner).
A tangent line has been added at an arbitrary point to illustrate the deviation of the curve from a straight line.
If we do negative binomial regression (on the unsmoothed data) then we get a better explanation for the data by allowing different generation times for the two variants: ΔAIC=7.8 for the period 2021-04-20 - 2021-06-20, and ΔAIC=47.1 for the period 2021-04-20 - 2021-07-08. That means that there is a significant deviation from being a straight line, and a difference of generation times is one possible way to explain this, or at least part of it.
Note that in the regression, there is a nuisance parameter for each time step that is optimised out, corresponding to factoring out the overall transmission rate (attributed to differing behaviour) that applies equally to both variants. Details in Appendix 2. Program used.
The outcome is that \[\lambda_t(\delta)-\rho\lambda_t(\alpha)\approx\lambda,\] for constants $$\rho$$ and $$\lambda$$, with \[\rho=1.137\; (1.046 - 1.229)\] using the time period 2021-04-20 - 2021-06-20, and \[\rho=1.249\; (1.155 - 1.343)\] using the time period 2021-04-20 - 2021-07-08.
The difference between this linear relation and the one above in the time period 2021-04-20 - 2021-05-31 is that this one covers a period where $$\lambda_t(\alpha)$$ and $$\lambda_t(\delta)$$ are non-constant. That raises the possibility of explaining $$\lambda_t(\delta)-\rho\lambda_t(\alpha)\approx\lambda$$ using the simple (constant distribution generation time) expression $$\lambda_t(\delta)T(\delta)-\lambda_t(\alpha)T(\alpha)=\log(R_t(\delta)/R_t(\alpha))$$, which would mean $$T(\alpha)/T(\delta)=\rho$$. In other words, one explanation for the bend in the curve in this graph is that Alpha has a generation time roughly 20% more than Delta. [Do error analysis.]
If we further assume that $$R_t(\delta)/R_t(\alpha)=1.65$$ as above, then we can combine the expressions and get estimates for the generation times separately. The point estimates using the time period 2021-04-20 - 2021-06-20 is $$T(\alpha)=5.0$$ days, and $$T(\delta)=4.4$$ days, which are in keeping with estimates that people have been using for the generation times. [Do error analysis.]
Non-constant generation time distribution
To do. Try different Gamma distributions, with the same means, for the generation times of Alpha and Delta, to see if different distribution shapes can explain the observations better than different means.
References
- https://www.gov.uk/government/publications/juniper-potential-community-transmission-of-b16172-inferred-by-s-gene-positivity-briefing-note-11-may-2021
- https://www.gov.uk/government/publications/investigation-of-novel-sars-cov-2-variant-variant-of-concern-20201201
- https://github.com/alex1770/Covid-19/tree/master/VOCgrowth/gentimetheory.2021-08-01