Difference between revisions of "UK variant comparison"
| Line 1: | Line 1: | ||
These graphs should auto-update every few days, according to when there is new sequence data from [https://www.cogconsortium.uk/tools-analysis/public-data-analysis-2/ COG-UK]. You may need to hit shift-reload or some such to defeat your browser's cache. | These graphs should auto-update every few days, according to when there is new sequence data from [https://www.cogconsortium.uk/tools-analysis/public-data-analysis-2/ COG-UK]. You may need to hit shift-reload or some such to defeat your browser's cache. | ||
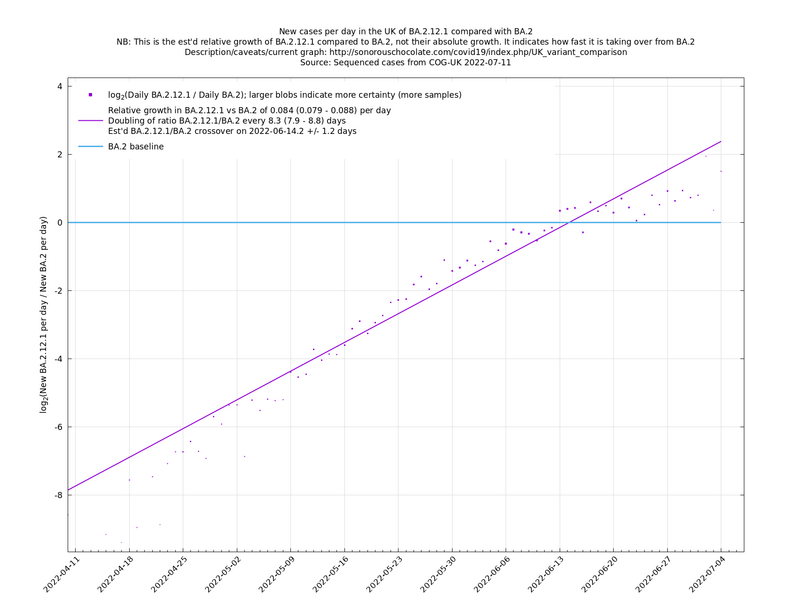
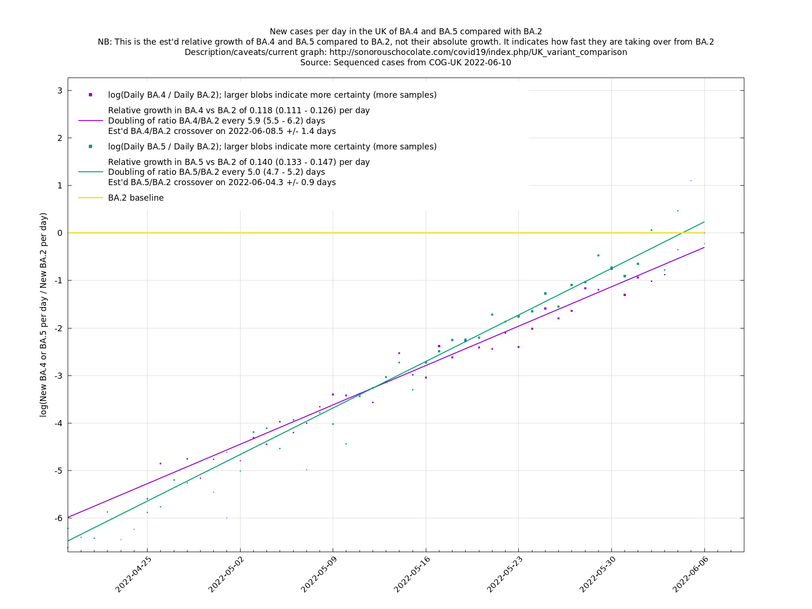
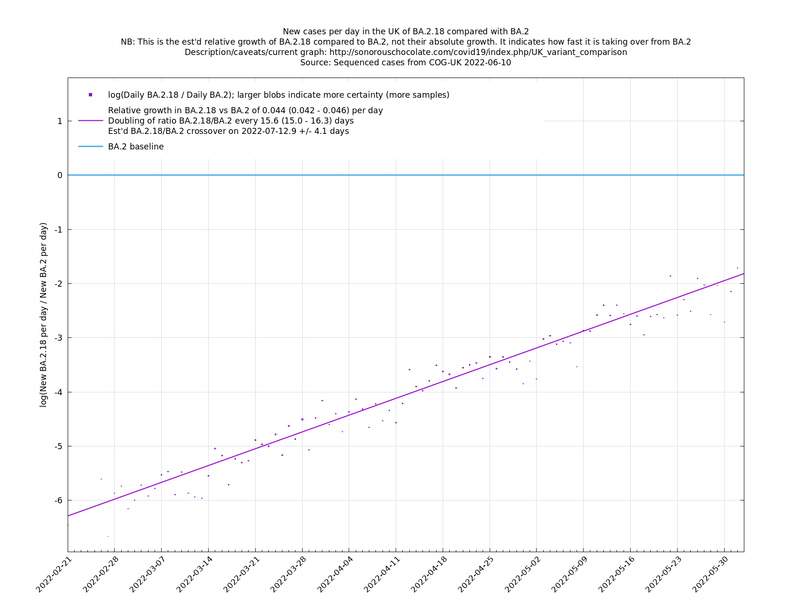
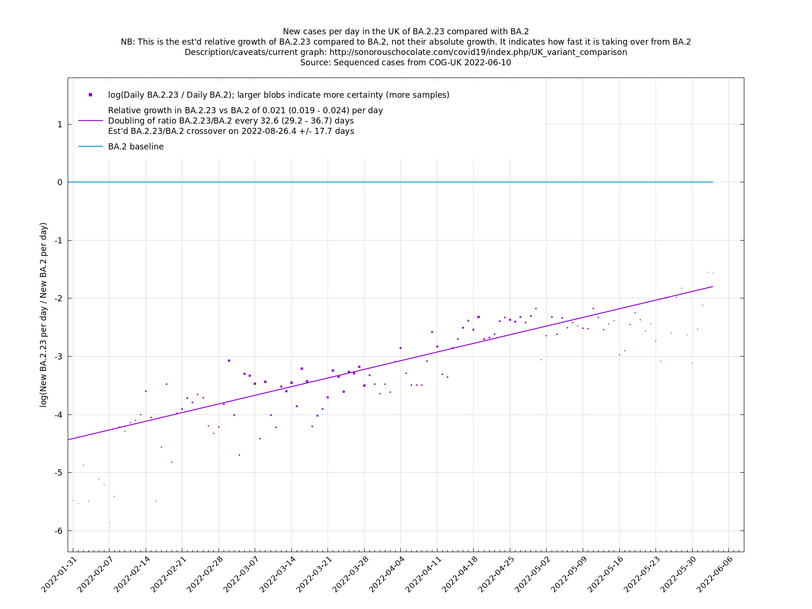
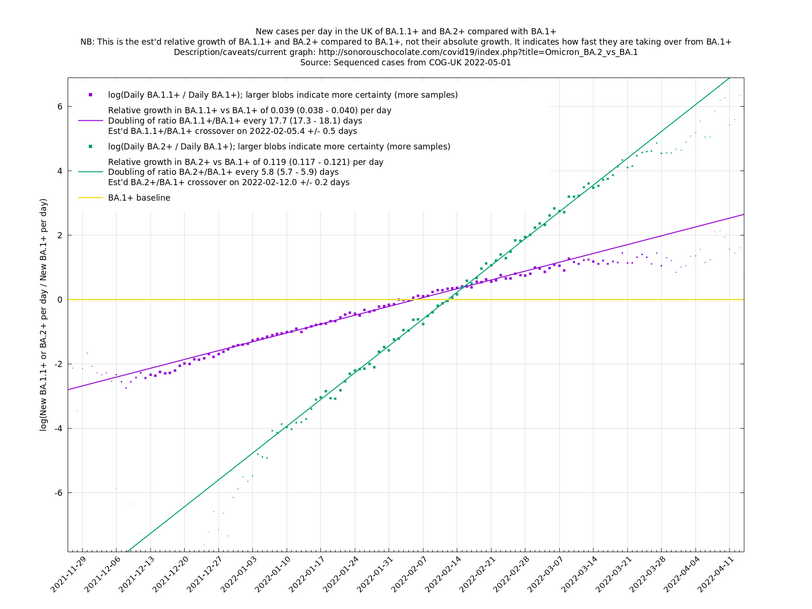
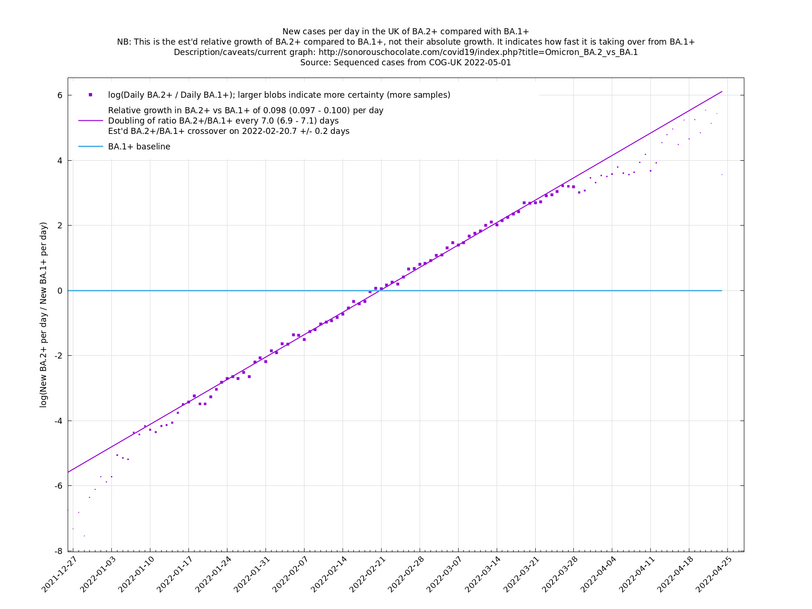
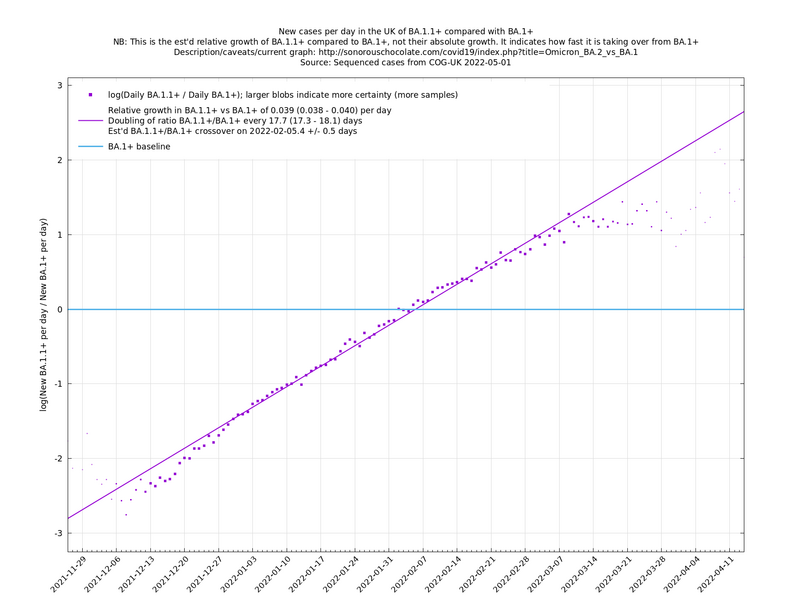
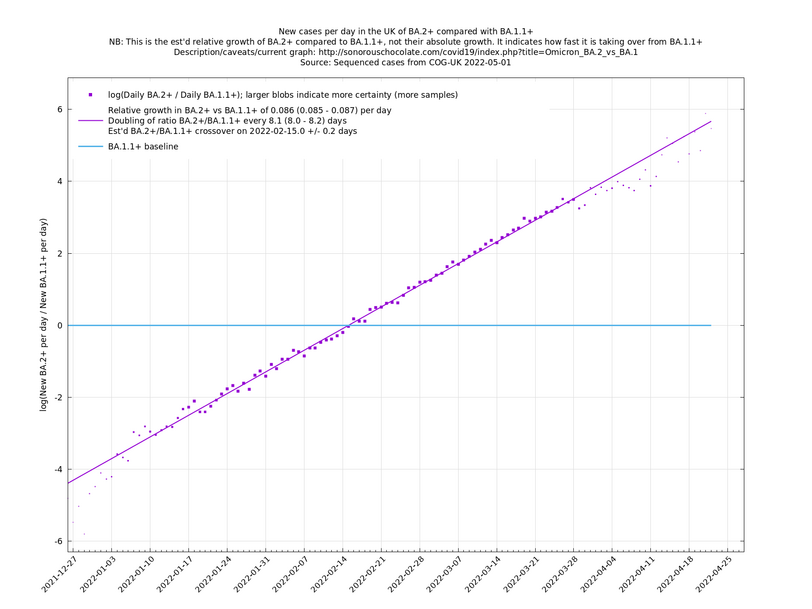
| − | These graphs compare one or more variants with another "baseline" variant. A y-co-ordinate of 0 corresponds to equal number of new cases per day of a variant compared to the baseline variant. The purpose of presenting it this way, with a particular chosen baseline variant, as opposed to | + | These graphs compare one or more variants with another "baseline" variant. A y-co-ordinate of 0 corresponds to equal number of new cases per day of a variant compared to the baseline variant. The purpose of presenting it this way, with a particular chosen baseline variant, as opposed to several variants on an equal footing, is that it means we expect to get straight lines to some reasonable approximation. |
Note that the growth estimates here rely on sequenced cases, of which there are currently about 3,500 per day, being representative of infections. This assumption may or may not hold, though it appears to work well in many cases. We often see a good linear fit over an extended period (say, several weeks) which arguably constitutes evidence in its favour, because many distortions you may think of, such as preferential sequencing of a particular variant for a limited time period, or different growth rates in different subpopulations, would often have the effect of knocking the relationship off being linear. | Note that the growth estimates here rely on sequenced cases, of which there are currently about 3,500 per day, being representative of infections. This assumption may or may not hold, though it appears to work well in many cases. We often see a good linear fit over an extended period (say, several weeks) which arguably constitutes evidence in its favour, because many distortions you may think of, such as preferential sequencing of a particular variant for a limited time period, or different growth rates in different subpopulations, would often have the effect of knocking the relationship off being linear. | ||
Revision as of 06:42, 16 May 2022
These graphs should auto-update every few days, according to when there is new sequence data from COG-UK. You may need to hit shift-reload or some such to defeat your browser's cache.
These graphs compare one or more variants with another "baseline" variant. A y-co-ordinate of 0 corresponds to equal number of new cases per day of a variant compared to the baseline variant. The purpose of presenting it this way, with a particular chosen baseline variant, as opposed to several variants on an equal footing, is that it means we expect to get straight lines to some reasonable approximation.
Note that the growth estimates here rely on sequenced cases, of which there are currently about 3,500 per day, being representative of infections. This assumption may or may not hold, though it appears to work well in many cases. We often see a good linear fit over an extended period (say, several weeks) which arguably constitutes evidence in its favour, because many distortions you may think of, such as preferential sequencing of a particular variant for a limited time period, or different growth rates in different subpopulations, would often have the effect of knocking the relationship off being linear.
Update 2022-05-14: Now including BA.4 and BA.5 vs BA.2. Note the large uncertainty in the growths of BA.2.12.1, BA.4 and BA.5. Removed XE because it was increasing too slowly to be of interest.
Update 2022-05-09: Now including BA.2.12.1 vs BA.2. (Treating BA.2.12 with S:L452Q as BA.2.12.1.)
Update 2022-05-05: Now more BA.2.18 and BA.2.23 have been classified, their growth advantage relative to BA.2 is seen to be more modest.
Update 2022-05-01: Now showing BA.2.18 and BA.2.23. These are both apparently showing a significant growth advantage against BA.2 (the currently dominant lineage), though this is based on low case numbers which means there is uncertainty as to whether these growth rates are correct or an early artefact.
Update 2022-04-07: The BA.* lineages are now classified into many more sublineages. For the purposes of these graphs, a suffix of '+' means that all sublineages are included along with the principal lineage. For example, BA.1+ includes BA.1 and BA.1.17. In the situation where a sublineage would be included under more than one of the lineages being compared, it is assigned to the most recent ancestor. For example in the graph of BA.1.1+ vs BA.1+, the sublineage BA.1.1.7 is assigned to BA.1.1+, not BA.1+. (The older graphs involving only BA.1+, BA.1.1+, BA.2+, all use the '+' suffix because they are continuations of graphs that began in January 2022 when BA.1, BA.1.1 and BA.2 weren't subdivided. Doing it this way allows you to see what was known in the past.)
Update 2022-04-07: Now including XE vs BA.2. The graph below suggests XE is only increasing very slowly relative to BA.2.
Program used for analysis: uk_var_comp.py.