Difference between revisions of "UK variant comparison"
| Line 1: | Line 1: | ||
| − | These graphs should auto-update every | + | = Self-updating graphs showing current situation = |
| + | |||
| + | These graphs should auto-update every day or two, according to when there is new sequence data from [https://www.cogconsortium.uk/tools-analysis/public-data-analysis-2/ COG-UK]. You may need to hit shift-reload or some such to defeat your browser's cache. | ||
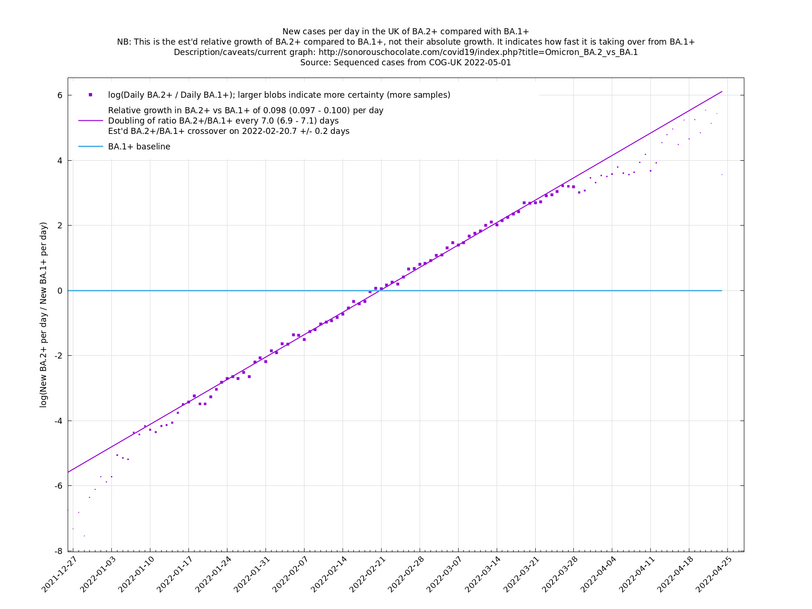
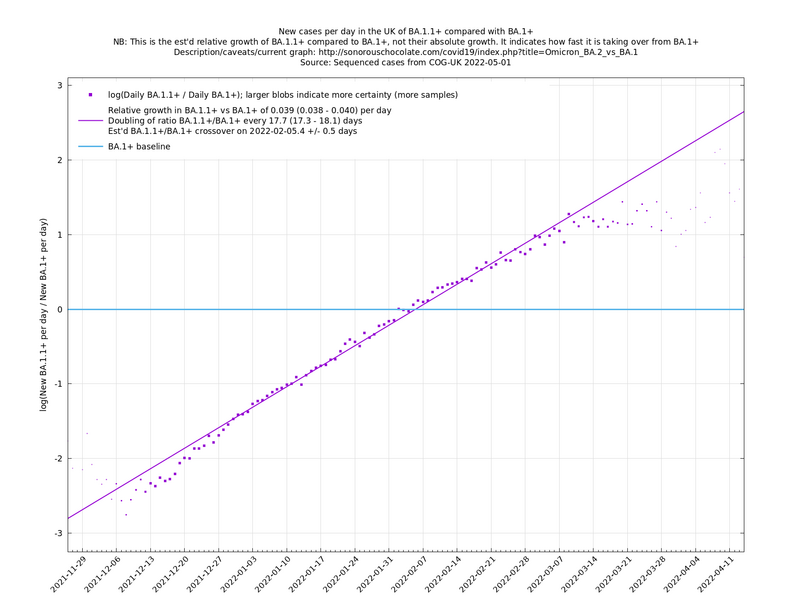
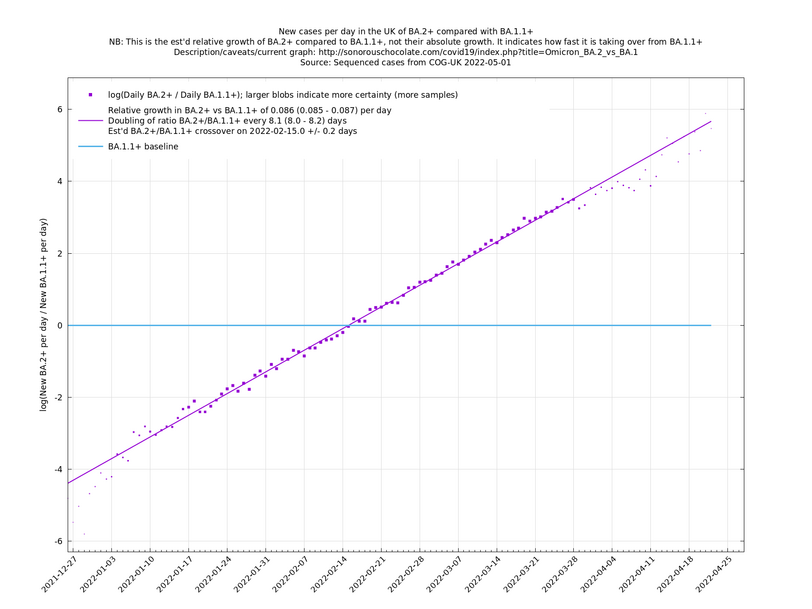
These graphs compare one or more variants with another "baseline" variant. A y-co-ordinate of 0 corresponds to equal number of new cases per day of a variant compared to the baseline variant. The purpose of presenting it this way, with a particular chosen baseline variant, as opposed to several variants on an equal footing, is that it means we expect to get straight lines to some reasonable approximation. | These graphs compare one or more variants with another "baseline" variant. A y-co-ordinate of 0 corresponds to equal number of new cases per day of a variant compared to the baseline variant. The purpose of presenting it this way, with a particular chosen baseline variant, as opposed to several variants on an equal footing, is that it means we expect to get straight lines to some reasonable approximation. | ||
| Line 5: | Line 7: | ||
Note that the growth estimates here rely on sequenced cases, of which there are currently about 1,000 per day, being representative of infections. This assumption may or may not hold, though it appears to work well in many cases. We often see a good linear fit over an extended period (say, several weeks) which arguably constitutes evidence in its favour, because many distortions you may think of, such as preferential sequencing of a particular variant for a limited time period, or different growth rates in different subpopulations, would often have the effect of knocking the relationship off being linear. | Note that the growth estimates here rely on sequenced cases, of which there are currently about 1,000 per day, being representative of infections. This assumption may or may not hold, though it appears to work well in many cases. We often see a good linear fit over an extended period (say, several weeks) which arguably constitutes evidence in its favour, because many distortions you may think of, such as preferential sequencing of a particular variant for a limited time period, or different growth rates in different subpopulations, would often have the effect of knocking the relationship off being linear. | ||
| − | + | Program used for analysis: [https://github.com/alex1770/Covid-19/blob/master/VOCgrowth/uk_var_comp.py uk_var_comp.py]. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
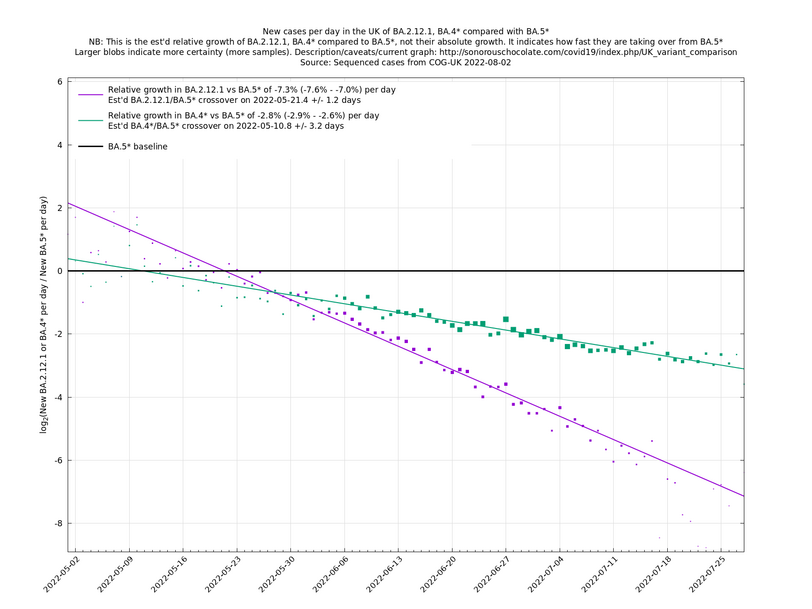
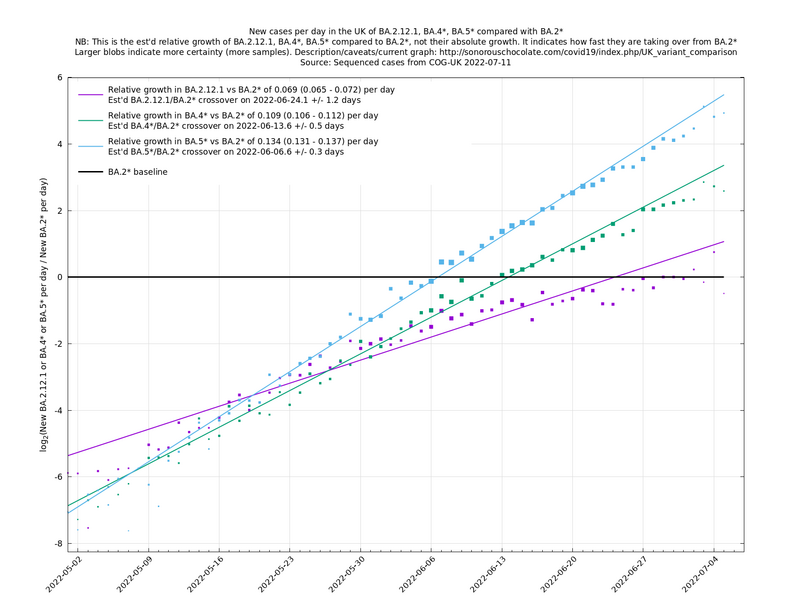
| − | + | As of 2022-07-11 in the UK, the overall view is that BA.5 and its subvariants, collectively written as BA.5*, are dominating, pulling clear of BA.4*, which in turn have prevailed over BA.2*. The strongest representative of BA.2* here is BA.2.12.1 (which has been strong in the US) and these three variant groups are shown in the following graph. | |
| − | + | [http://sonorouschocolate.com/covid19/extdata/UK_BA.5*_BA.2.12.1_BA.4*.png <img src="http://sonorouschocolate.com/covid19/extdata/UK_BA.5*_BA.2.12.1_BA.4*.small.png">] | |
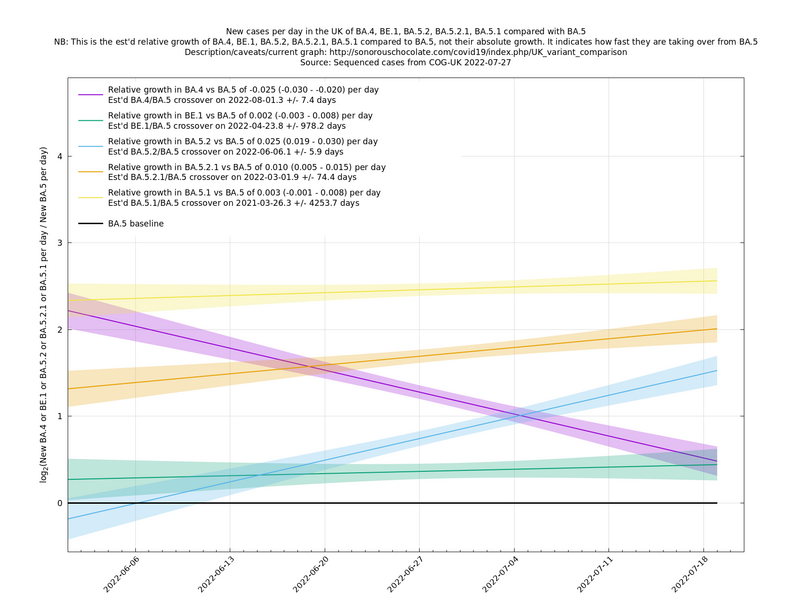
| − | + | BA.5 has evolved some subvariants which are showing a small but significant growth advantage over vanilla BA.5. The strongest subvariants, together with BA.4, are shown in the following graph, BE.1 being an alias for BA.5.3.1.1 (daily blobs removed for clarity). These extra growth advantages (of up to 3%) over BA.5 could be a nuisance, especially if they represent immune escape rather than transmissibility, but to some extent this is already priced in to BA.5*'s growth and there appear currently to be no other variants imminently threatening to take over from BA.5* in the UK, so when the current summer wave subsides it seems likely we will have at least a couple of months of respite from Covid-19 waves. Further ahead than that is hard to predict because it relies on knowing how future variants evolve and behave. | |
| − | [http://sonorouschocolate.com/covid19/extdata/UK_BA.2_BA.2. | + | [http://sonorouschocolate.com/covid19/extdata/UK_BA.5_BA.4_BE.1_BA.5.2_BA.5.2.1_BA.5.1.png <img src="http://sonorouschocolate.com/covid19/extdata/UK_BA.5_BA.4_BE.1_BA.5.2_BA.5.2.1_BA.5.1.small.png">] |
| − | |||
| − | |||
| − | |||
| − | |||
= Graphs below featuring extinct or disappearing baseline lineages are no longer updated = | = Graphs below featuring extinct or disappearing baseline lineages are no longer updated = | ||
Revision as of 10:49, 11 July 2022
Self-updating graphs showing current situation
These graphs should auto-update every day or two, according to when there is new sequence data from COG-UK. You may need to hit shift-reload or some such to defeat your browser's cache.
These graphs compare one or more variants with another "baseline" variant. A y-co-ordinate of 0 corresponds to equal number of new cases per day of a variant compared to the baseline variant. The purpose of presenting it this way, with a particular chosen baseline variant, as opposed to several variants on an equal footing, is that it means we expect to get straight lines to some reasonable approximation.
Note that the growth estimates here rely on sequenced cases, of which there are currently about 1,000 per day, being representative of infections. This assumption may or may not hold, though it appears to work well in many cases. We often see a good linear fit over an extended period (say, several weeks) which arguably constitutes evidence in its favour, because many distortions you may think of, such as preferential sequencing of a particular variant for a limited time period, or different growth rates in different subpopulations, would often have the effect of knocking the relationship off being linear.
Program used for analysis: uk_var_comp.py.
As of 2022-07-11 in the UK, the overall view is that BA.5 and its subvariants, collectively written as BA.5*, are dominating, pulling clear of BA.4*, which in turn have prevailed over BA.2*. The strongest representative of BA.2* here is BA.2.12.1 (which has been strong in the US) and these three variant groups are shown in the following graph.
BA.5 has evolved some subvariants which are showing a small but significant growth advantage over vanilla BA.5. The strongest subvariants, together with BA.4, are shown in the following graph, BE.1 being an alias for BA.5.3.1.1 (daily blobs removed for clarity). These extra growth advantages (of up to 3%) over BA.5 could be a nuisance, especially if they represent immune escape rather than transmissibility, but to some extent this is already priced in to BA.5*'s growth and there appear currently to be no other variants imminently threatening to take over from BA.5* in the UK, so when the current summer wave subsides it seems likely we will have at least a couple of months of respite from Covid-19 waves. Further ahead than that is hard to predict because it relies on knowing how future variants evolve and behave.
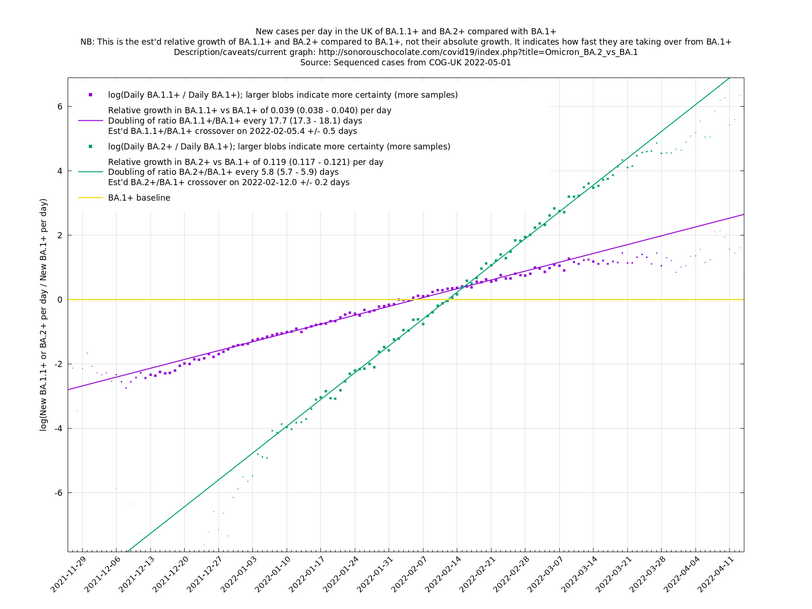
Graphs below featuring extinct or disappearing baseline lineages are no longer updated
(Note the y-scales below use natural logs.)